5 Differential Binding Region - DiffBind
Based on 71231 consensus peaks (see here)
Software package used: DiffBind
Background normalisationmethod usingDiffBind3
li.diff<-list()
li.diff[['bg']][["female"]]<-dba(dbo.cnt,dbo.cnt$masks$F) # isa DBA
li.diff[['bg']][["male"]]<- dba(dbo.cnt,dbo.cnt$masks$M) # isa DBA
li.diff[['bg']]<-lapply(li.diff[['bg']], dba.normalize, method=DBA_ALL_METHODS,normalize=DBA_NORM_NATIVE,library=DBA_LIBSIZE_FULL,background=TRUE)
li.diff[['bg']]<-lapply(li.diff[['bg']], dba.contrast, design="~Replicate+Factor")
li.diff[['bg']]<-lapply(li.diff[['bg']], dba.analyze, method=DBA_ALL_METHODS)
lapply(li.diff[['bg']], dba.show, bContrasts=TRUE)
lapply(li.diff[['bg']][c('male')], dba.report, method=DBA_ALL_METHODS,bDB=T,bGain=T,bLoss=T)
lapply(li.diff[['bg']], dba.normalize, bRetrieve=T,method=DBA_ALL_METHODS)[["female"]]5.1 No. of DBR
5.1.1 adjutedd P-val<0.05
| Factor | Group | Samples | Group2 | Samples2 | DB.edgeR | DB.DESeq2 |
|---|---|---|---|---|---|---|
| Factor | FD | 5 | FV | 5 | 0 | 0 |
| Factor | MD | 6 | MV | 6 | 213 | 147 |
lapply(li.diff[['bg']][c('male')], dba.report, method=DBA_ALL_METHODS,bDB=T,bGain=T,bLoss=T)## $male
## 6 Samples, 246 sites in matrix:
## Contrast Direction DB Method Intervals
## 1 MD vs. MV All DB edgeR 213
## 2 MD vs. MV Gain DB edgeR 21
## 3 MD vs. MV Loss DB edgeR 192
## 4 MD vs. MV All DB DESeq2 147
## 5 MD vs. MV Gain DB DESeq2 10
## 6 MD vs. MV Loss DB DESeq2 137## $male
## 2 Samples, 246 sites in matrix:
## Contrast Direction DB Method Intervals
## 1 MD vs. MV All DB edgeR 213
## 2 MD vs. MV All DB DESeq2 147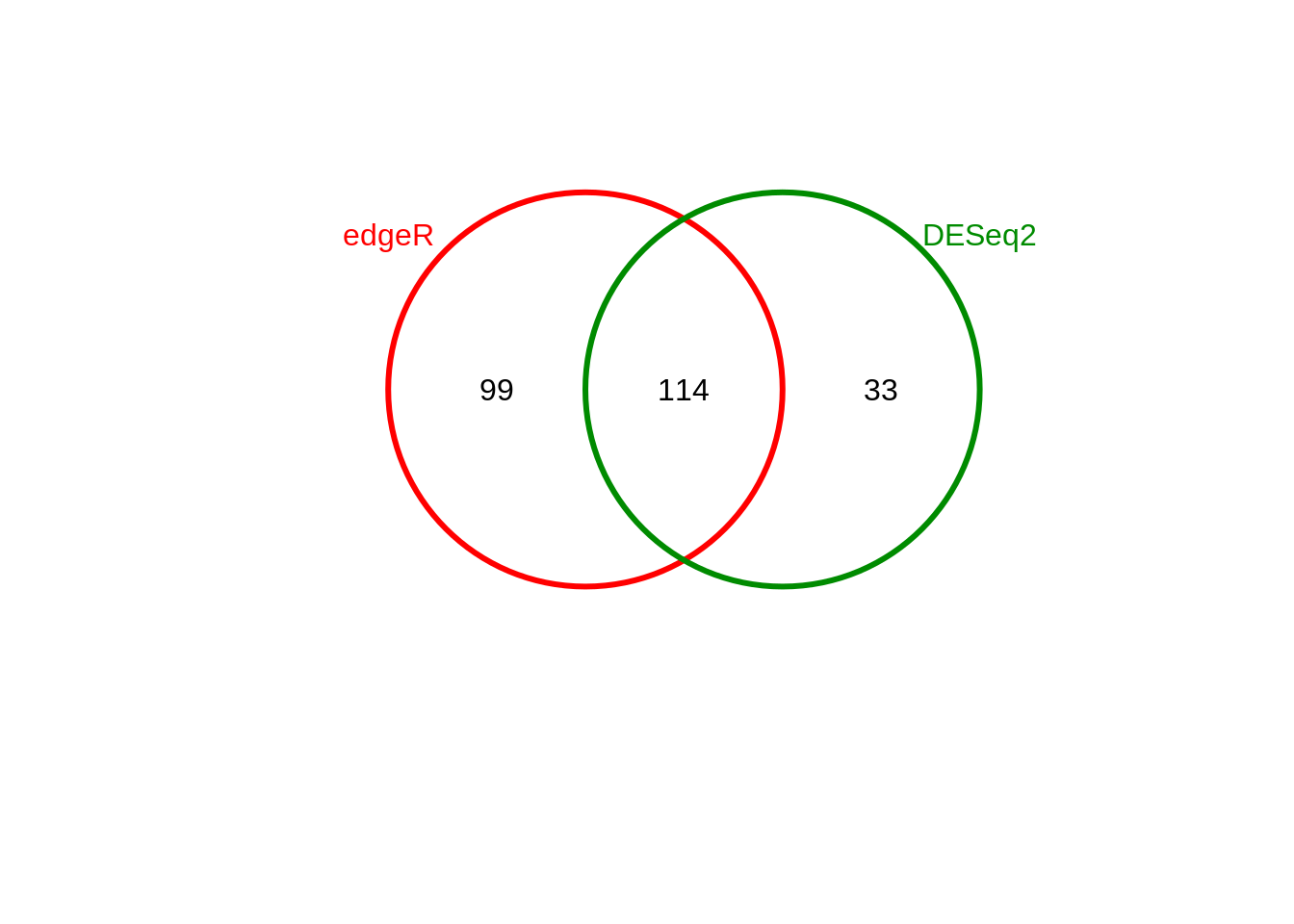
Figure 5.1: DBR between edgeR & DESeq2 (MD vs MV)
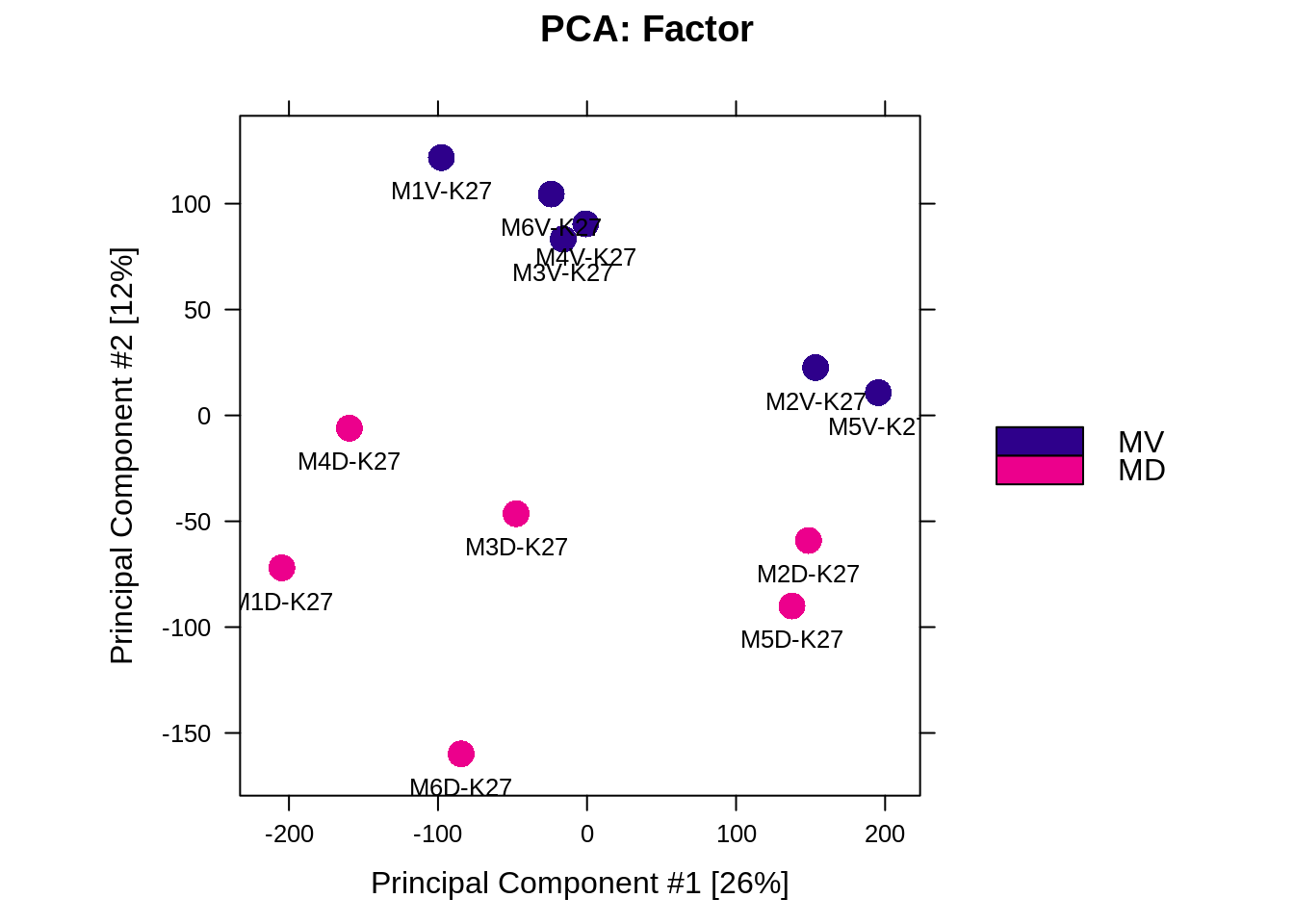
Figure 5.2: PCA based on DBR (MD vs MV)
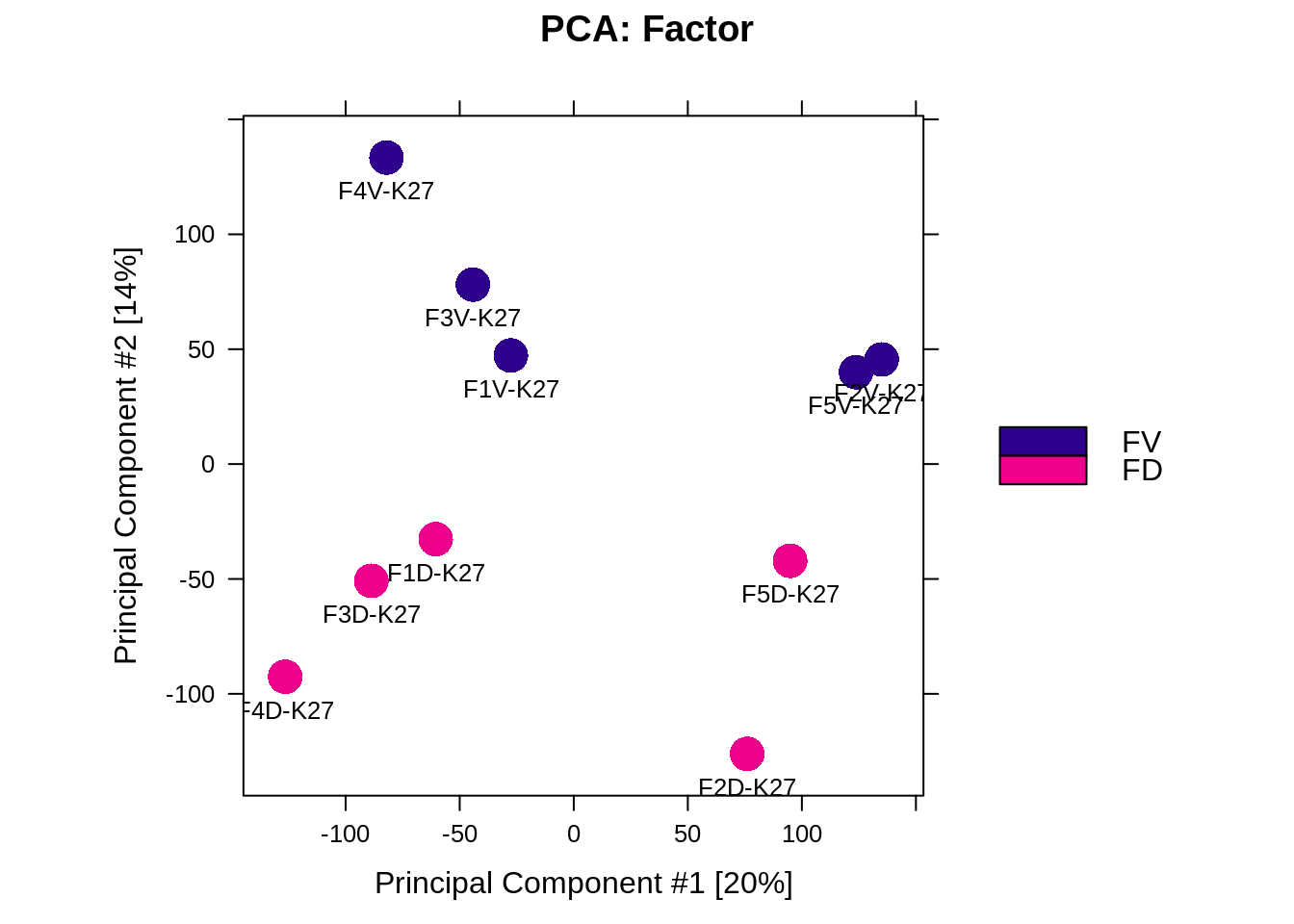
Figure 5.3: PCA based on DBR (FD vs FV)
5.1.2 adjutedd P-val<0.01:
| Factor | Group | Samples | Group2 | Samples2 | DB.edgeR | DB.DESeq2 |
|---|---|---|---|---|---|---|
| Factor | FD | 5 | FV | 5 | 0 | 0 |
| Factor | MD | 6 | MV | 6 | 51 | 28 |
5.2 Genomic feature of DBRs
| Feature | DESeq2 | edgeR |
|---|---|---|
| Promoter (<=1kb) | 24.489796 | 30.516432 |
| Promoter (1-2kb) | 6.802721 | 9.859155 |
| 3’ UTR | 5.442177 | 4.225352 |
| 1st Exon | 2.040816 | 1.877934 |
| Other Exon | 2.721088 | 3.755869 |
| 1st Intron | 10.204082 | 7.981221 |
| Other Intron | 17.687075 | 15.962441 |
| Downstream (<=300) | 2.040816 | 1.408451 |
| Distal Intergenic | 28.571429 | 24.413145 |
| Feature | DESeq2 | edgeR |
|---|---|---|
| Promoter (<=1kb) | 36 | 65 |
| Promoter (1-2kb) | 10 | 21 |
| 3’ UTR | 8 | 9 |
| 1st Exon | 3 | 4 |
| Other Exon | 4 | 8 |
| 1st Intron | 15 | 17 |
| Other Intron | 26 | 34 |
| Downstream (<=300) | 3 | 3 |
| Distal Intergenic | 42 | 52 |
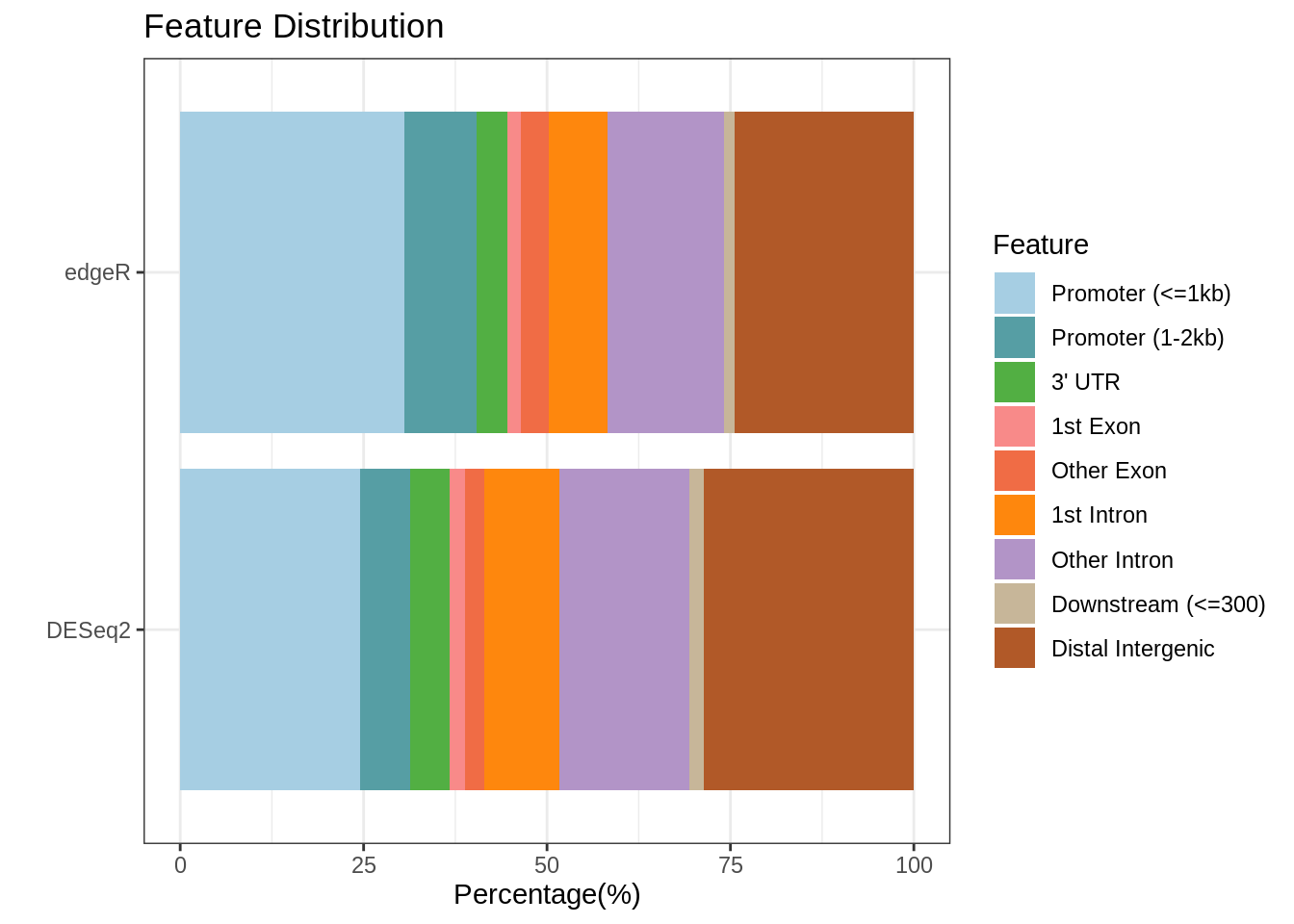
Figure 5.4: Genomic feautres of peaks
5.3 DBR from DESeq2
- Unfiltered list of DBR: MD vs MF
5.3.1 Normalisation factors
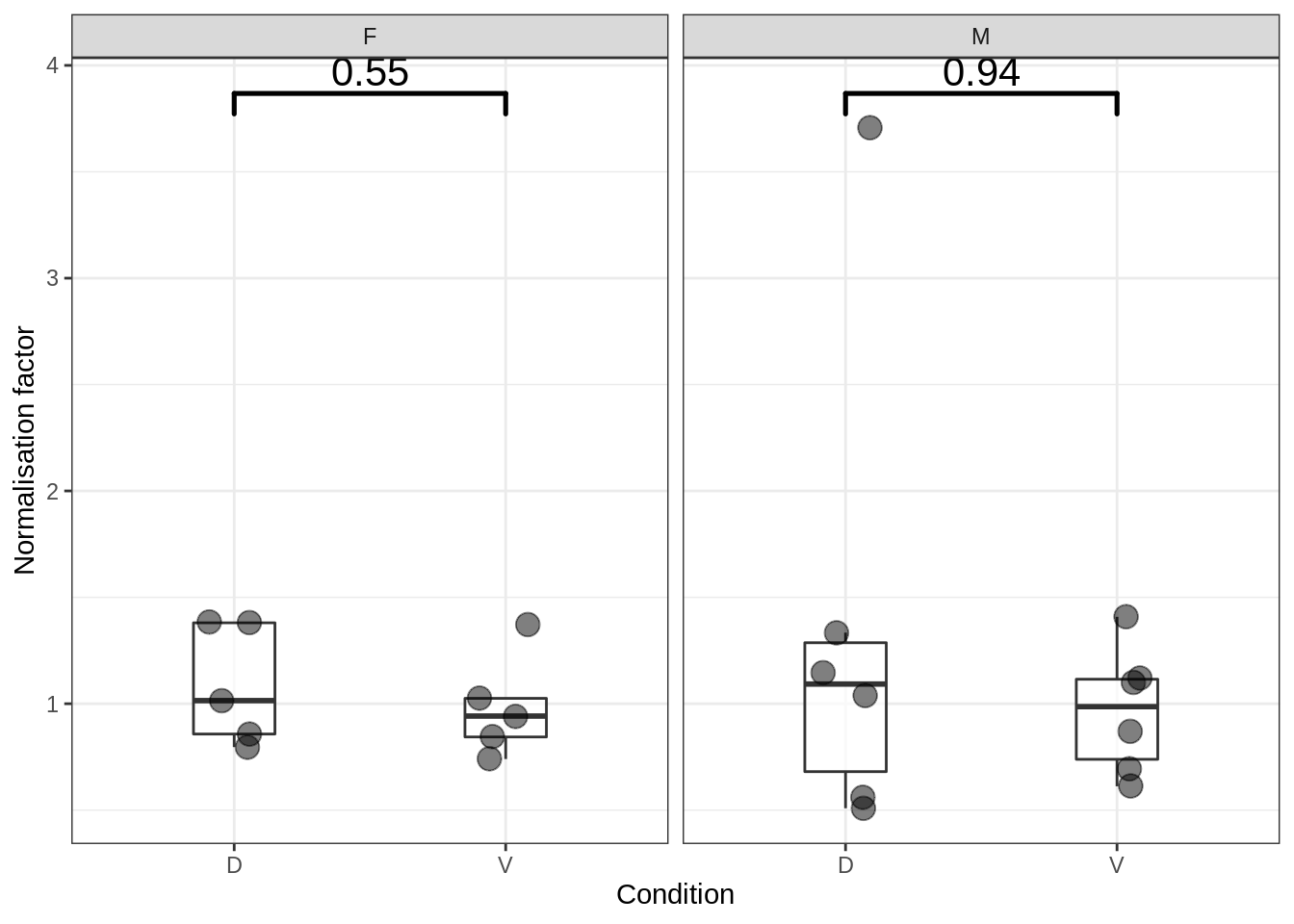
Figure 5.5: Normalisation factors by group
5.3.2 Male (MD vs MV)
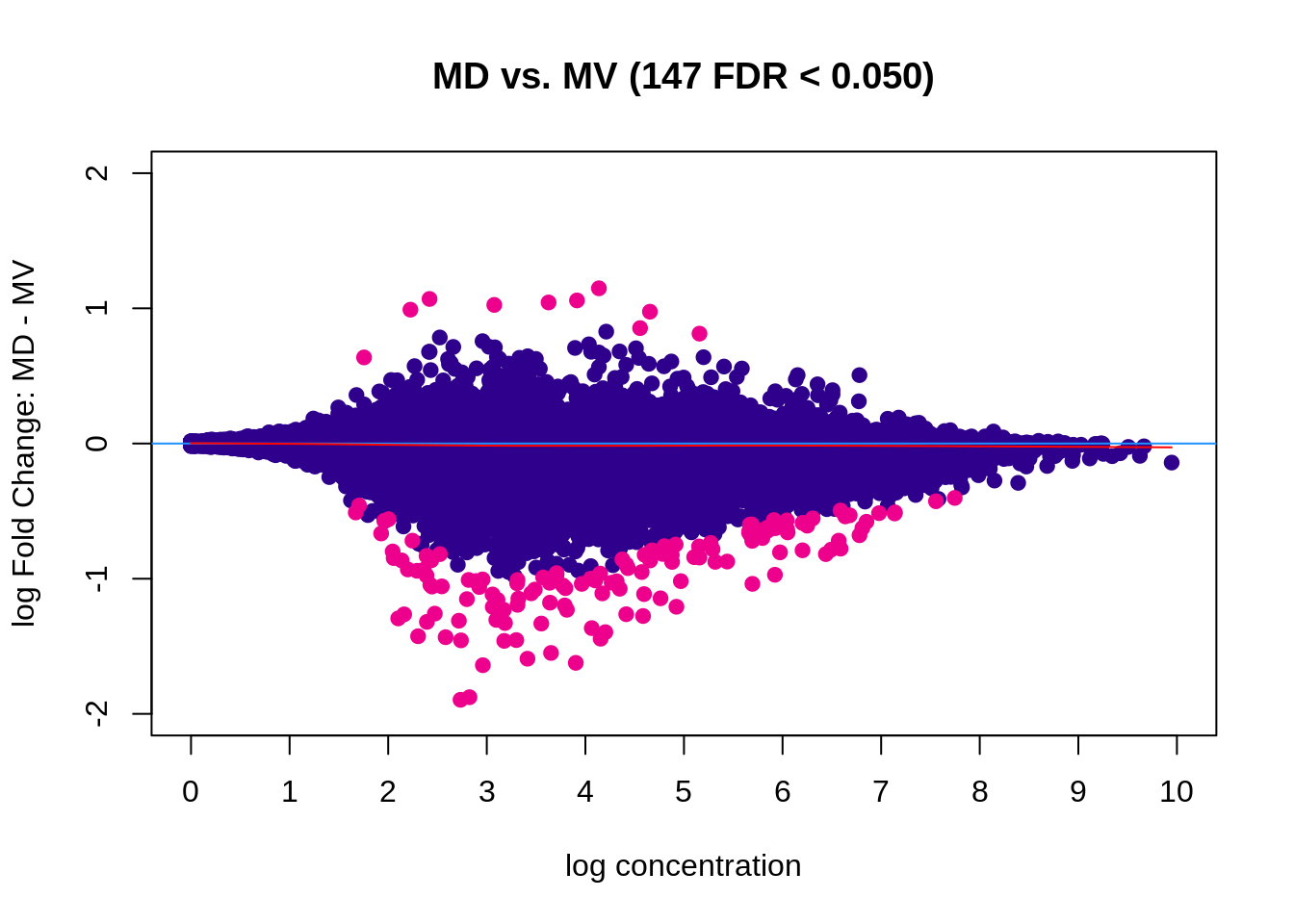
Figure 5.6: MA plot (DESeq2)
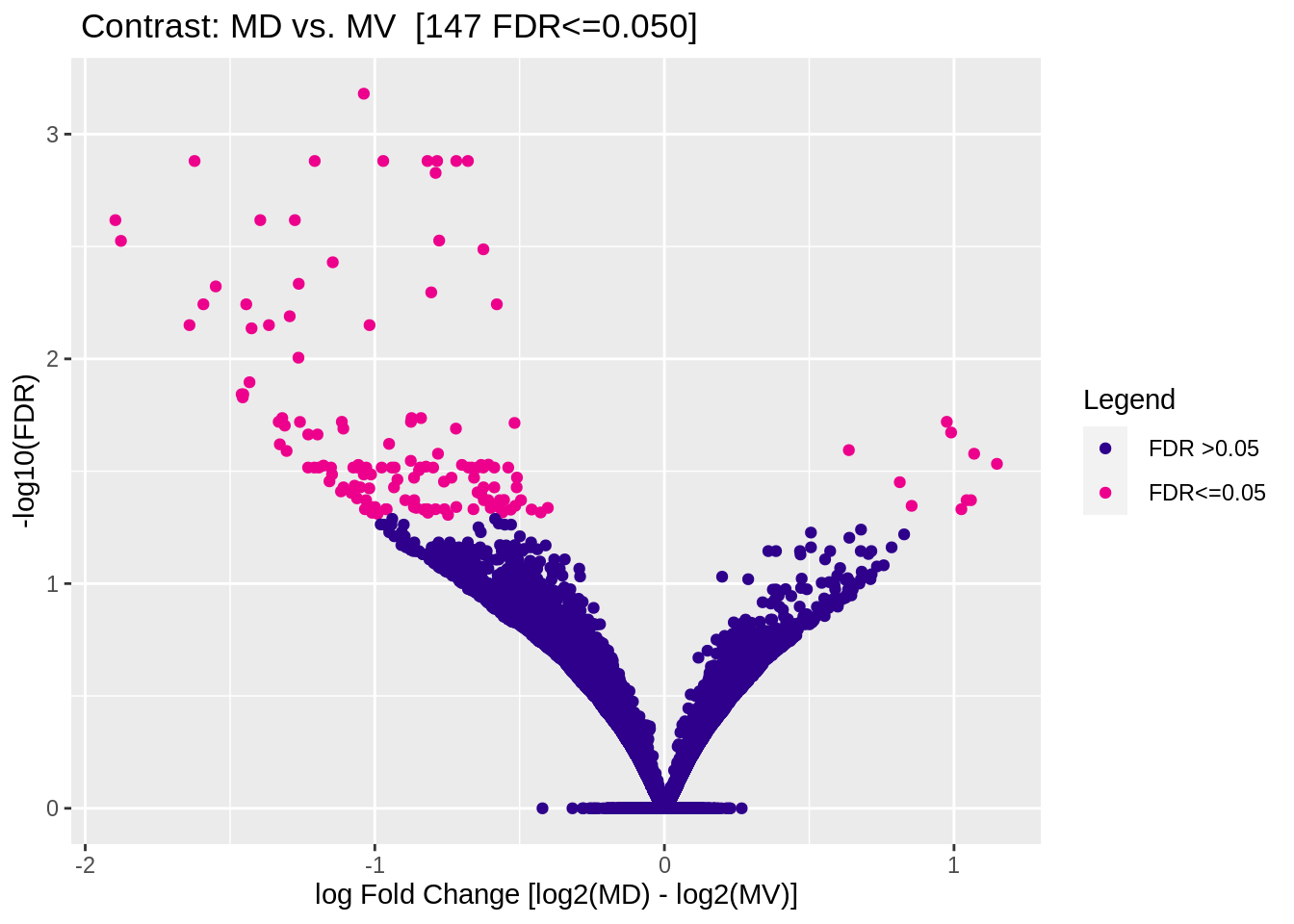
Figure 5.7: Volcano plot (DESeq2)
5.4 DBR from edgeR
- Unfiltered list of DBR: MD vs MF
5.4.1 Normalisation factors
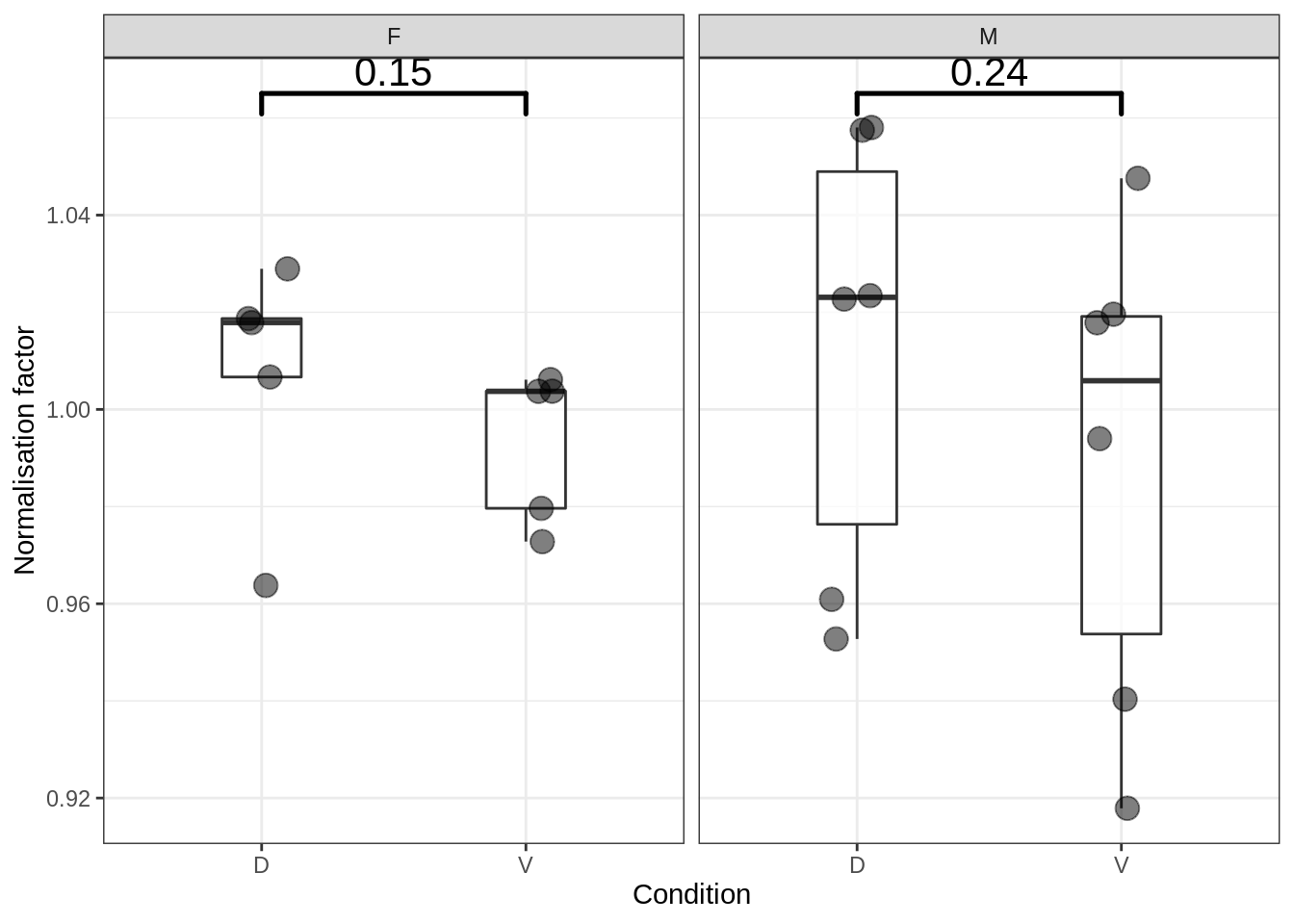
Figure 5.8: Normalisation factors by group
5.4.2 Male (MD vs MV)
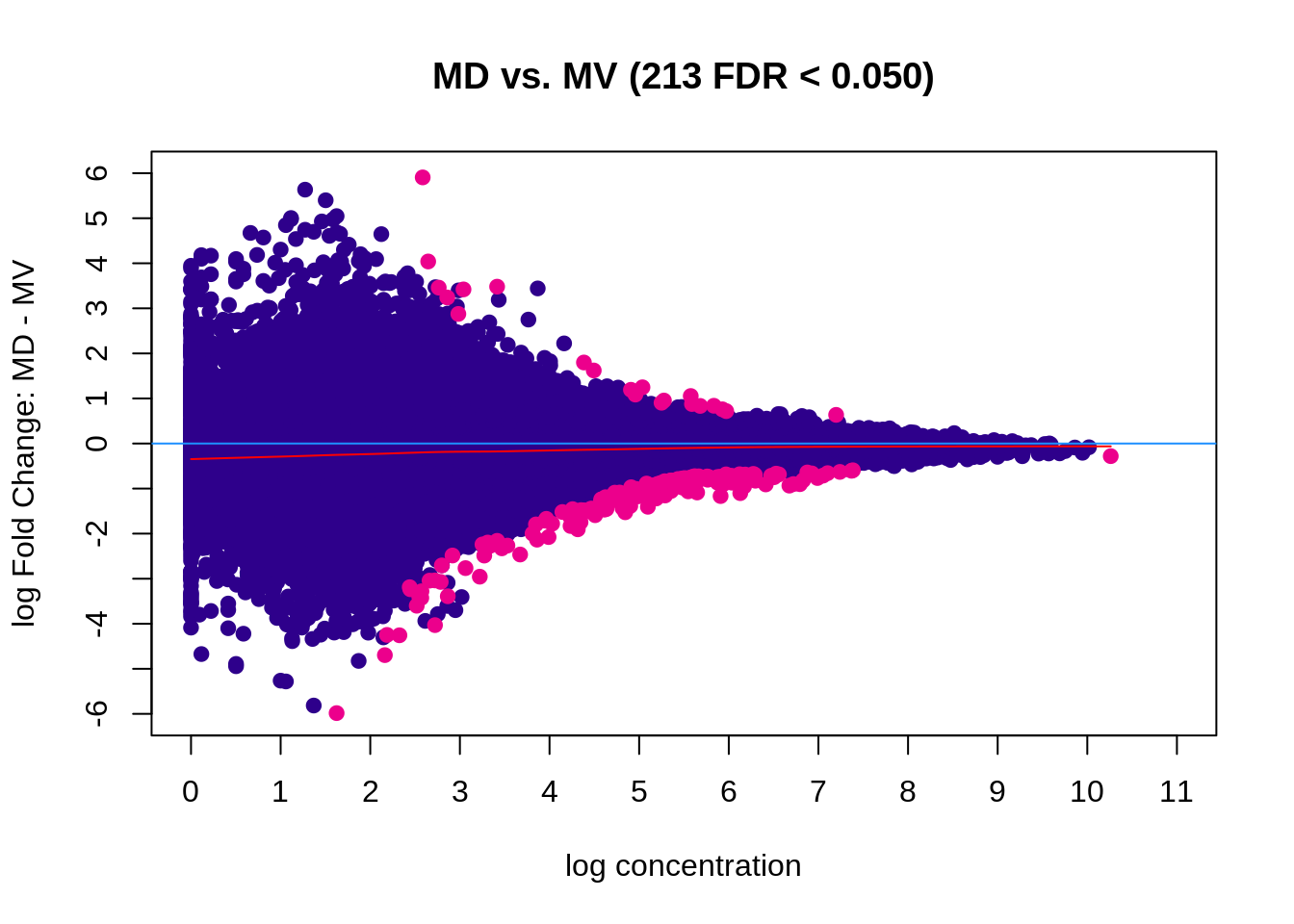
Figure 5.9: MA plot (edgeR)
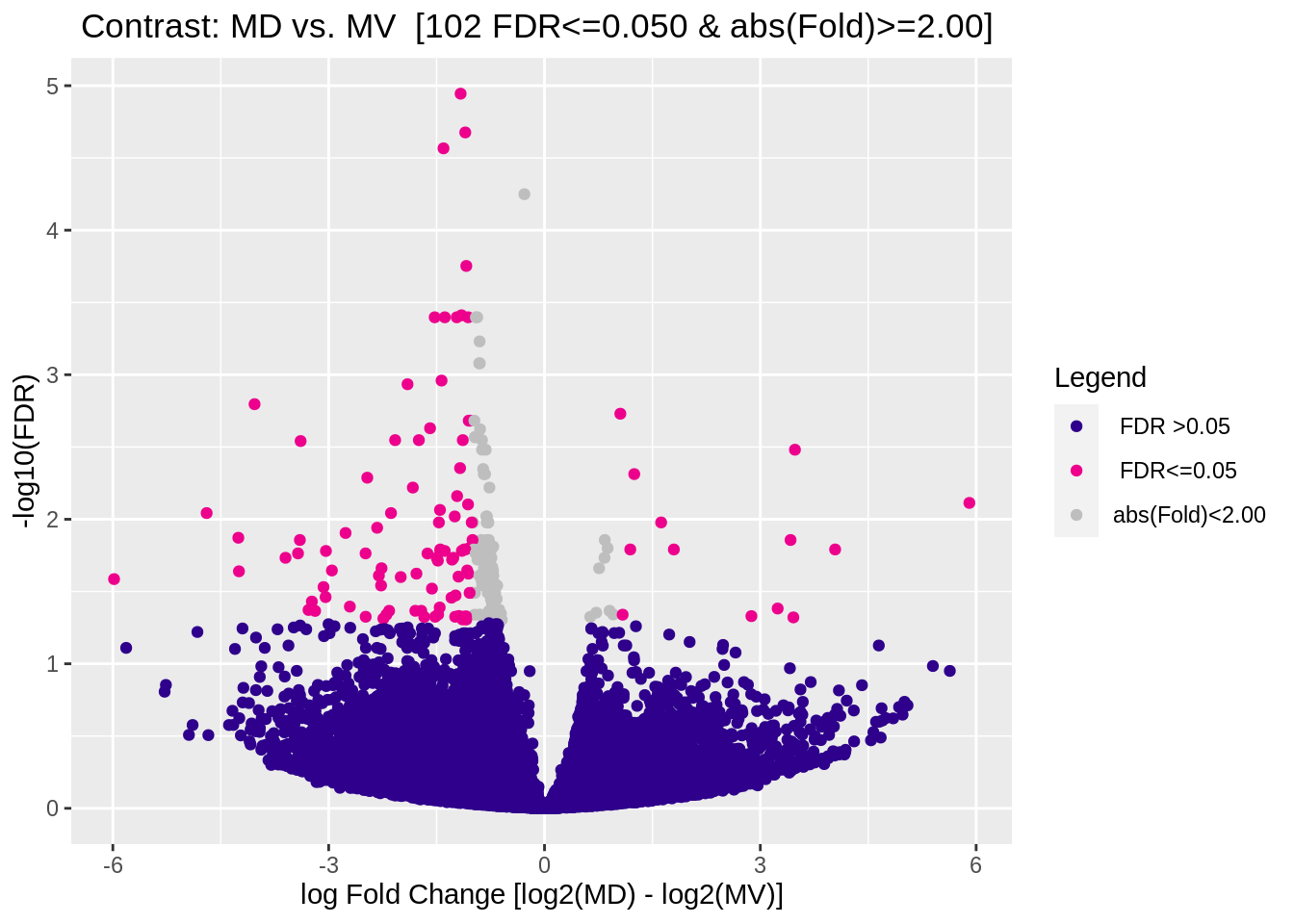
Figure 5.10: Volcano plot (edgeR)
5.5 Normalisation parameters
lapply(li.diff[['bg']], dba.normalize, bRetrieve=T,method=DBA_ALL_METHODS)## $female
## $female$edgeR
## $female$edgeR$background
## [1] TRUE
##
## $female$edgeR$norm.method
## [1] "TMM"
##
## $female$edgeR$norm.factors
## [1] 1.0037253 1.0178863 0.9796488 1.0187121 1.0037716 1.0066757 1.0061241 1.0289548 0.9727954 0.9637627
##
## $female$edgeR$lib.method
## [1] "background"
##
## $female$edgeR$lib.sizes
## [1] 14406659 14313568 9052547 8273839 10782259 10634010 7783031 8816437 10163025 15059456
##
## $female$edgeR$control.subtract
## [1] TRUE
##
## $female$edgeR$filter.value
## [1] 1
##
##
## $female$DESeq2
## $female$DESeq2$background
## [1] TRUE
##
## $female$DESeq2$norm.method
## [1] "RLE"
##
## $female$DESeq2$norm.factors
## F1V-K27 F1D-K27 F2V-K27 F2D-K27 F3V-K27 F3D-K27 F4V-K27 F4D-K27 F5V-K27 F5D-K27
## 1.3711165 1.3802589 0.8437394 0.7962241 1.0251811 1.0144064 0.7400174 0.8575304 0.9419272 1.3851355
##
## $female$DESeq2$lib.method
## [1] "background"
##
## $female$DESeq2$lib.sizes
## [1] 14406659 14313568 9052547 8273839 10782259 10634010 7783031 8816437 10163025 15059456
##
## $female$DESeq2$control.subtract
## [1] TRUE
##
## $female$DESeq2$filter.value
## [1] 1
##
##
## $female$background
## $female$background$binned
## class: RangedSummarizedExperiment
## dim: 188762 10
## metadata(6): spacing width ... param final.ext
## assays(1): counts
## rownames: NULL
## rowData names(0):
## colnames(10): F1V-K27 F1D-K27 ... F5V-K27 F5D-K27
## colData names(4): bam.files totals ext rlen
##
## $female$background$bin.size
## [1] 15000
##
## $female$background$back.calc
## [1] "Background bins"
##
##
##
## $male
## $male$edgeR
## $male$edgeR$background
## [1] TRUE
##
## $male$edgeR$norm.method
## [1] "TMM"
##
## $male$edgeR$norm.factors
## [1] 1.0475833 1.0580352 0.9403566 0.9527507 1.0195866 1.0227243 0.9939970 1.0574761 0.9178885 0.9608961 1.0178277
## [12] 1.0234355
##
## $male$edgeR$lib.method
## [1] "background"
##
## $male$edgeR$lib.sizes
## [1] 7842803 6256923 7620403 45662054 16275364 12003441 13043263 14752067 14301167 13951545 10101914 5878181
##
## $male$edgeR$control.subtract
## [1] TRUE
##
## $male$edgeR$filter.value
## [1] 1
##
##
## $male$DESeq2
## $male$DESeq2$background
## [1] TRUE
##
## $male$DESeq2$norm.method
## [1] "RLE"
##
## $male$DESeq2$norm.factors
## M1V-K27 M1D-K27 M2V-K27 M2D-K27 M3V-K27 M3D-K27 M4V-K27 M4D-K27 M5V-K27 M5D-K27 M6V-K27
## 0.6945813 0.5609957 0.6119628 3.7079627 1.4081663 1.0398274 1.1014219 1.3339729 1.1196840 1.1447977 0.8711595
## M6D-K27
## 0.5085670
##
## $male$DESeq2$lib.method
## [1] "background"
##
## $male$DESeq2$lib.sizes
## [1] 7842803 6256923 7620403 45662054 16275364 12003441 13043263 14752067 14301167 13951545 10101914 5878181
##
## $male$DESeq2$control.subtract
## [1] TRUE
##
## $male$DESeq2$filter.value
## [1] 1
##
##
## $male$background
## $male$background$binned
## class: RangedSummarizedExperiment
## dim: 189298 12
## metadata(6): spacing width ... param final.ext
## assays(1): counts
## rownames: NULL
## rowData names(0):
## colnames(12): M1V-K27 M1D-K27 ... M6V-K27 M6D-K27
## colData names(4): bam.files totals ext rlen
##
## $male$background$bin.size
## [1] 15000
##
## $male$background$back.calc
## [1] "Background bins"